Get in touch
555-555-5555
mymail@mailservice.com
Culturing Chlamydomonas reinhardtii - ALG App008
algenuity • Aug 16, 2016
Background
Chlamydomonas reinhardtii
is a motile, unicellular green microalga typically measuring around 10 µm in diameter. It is widely distributed, and is often isolated from soil and freshwater samples. C. reinhardtii
has been used as a model organism for over 70 years for both basic and applied research, largely due to its ease of cultivation, rapid doubling time of 6-8 h, and established molecular toolbox (Harris, 2009). Noted areas of study include photosynthesis, phototaxis, cell wall biogenesis, cell cycle events, flagella assembly, mating processes, and nuclear/chloroplast interactions (Rochaix, 1995; Shimogawara et al., 1998; Merchant et al., 2007). Annotated sequences are available for the nuclear, chloroplast and mitochondrial genomes (Merchant et al., 2007, Scaife et al., 2015), and several extensive libraries of mutants have been generated. Transformation of all three genomes have been demonstrated, with nuclear and chloroplast manipulation becoming routine (Boynton et al. 1988; Kindle et al. 1989; Sodeinde and Kindle. 1993).
Recently C. reinhardtii
has gained attention as a platform for commercial applications; these include recombinant protein expression (Mayfield et al., 2007; Rosales-Mendoza et al., 2012), biohydrogen production (Torzillo et al., 2015), and as a model testbed for biofuel technologies prior to shuttling into more industrially relevant, but less easy to manipulate, biofuel production strains.
Cultivation of C. reinhardtii
is typically conducted mixotrophically on TAP (tris acetate phosphate) medium. Although suitable for lab-scale work, TAP medium is not appropriate for scale up due to its relatively high cost and susceptibility to contamination.
Aim
To compare the growth of C. reinhardtii
CC1690 under mixotrophic (TAP medium) and phototrophic (HSM medium) conditions in the Algem photobioreactor.
Experimental Design
Two starter cultures of C. reinhardtii
CC1690 were inoculated from plates in either TAP or HSM. Cultures were incubated at 20°C, with 40 µE/m²/s continuous light, and 120 rpm agitation (Innova 44, New Brunswick Scientific, New Jersey) for four days, then sub-cultured. Starter cultures in mid-logarithmic phase were used to inoculate duplicate 400 ml cultures of HSM and TAP medium which were then cultured in Algems at 25°C, with 200 µE/m²/s continuous light (85: 15 white: red mix), at 100 rpm agitation and 10cc/minute aeration with 5 % CO₂for a period of five days.
Results
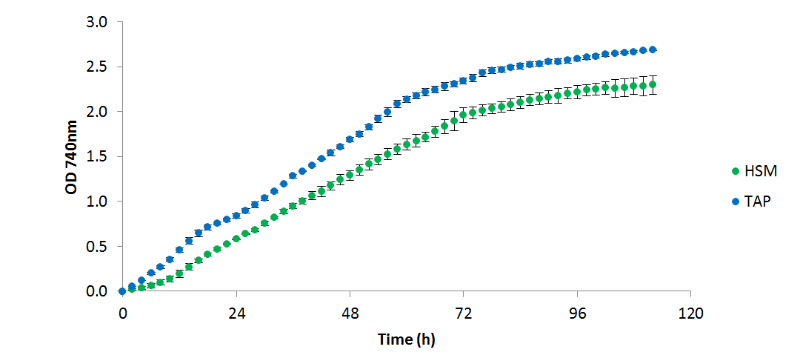
Figure 1 - C. reinhardtii
CC 1690 cultured in HSM and TAP medium at 25°C, 200 µE/m²/s continuous light (85: 15 white; red mix), 100 rpm, with 10 cm³/min aeration with 5 % CO₂, (n=2)
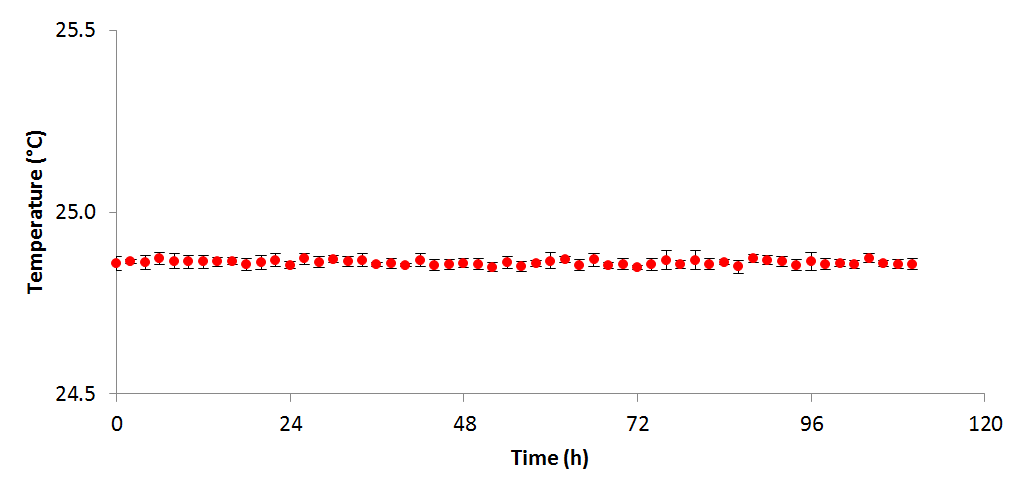
Figure 2 - Temperature variation over the course of the above experiment (n=4)
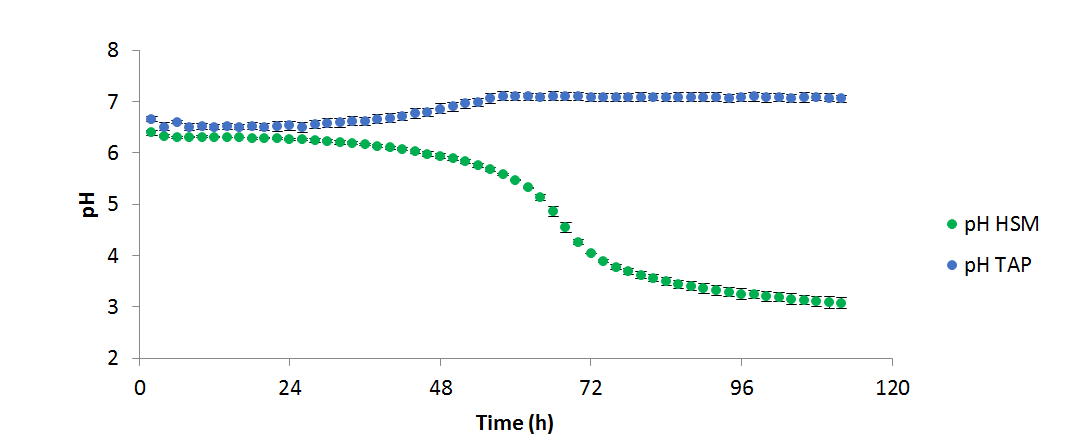
Figure 3 - pH variation over the course of the above experiment (n=2)
Discussion
It was observed that C. reinhardtii
grew more productively in TAP than HSM; however, it is unclear from these data whether this is due to the available carbon source (acetate and 5% CO₂vs. 5% CO₂only), or the buffering effects of acetate consumption. When algal cells are grown on NH₄+ a drop in pH is generally seen, as the ammonium symporter pumps out a proton into the medium for each ammonium ion transported into the cell. Conversely, when acetate is consumed, the pH increases as the tris: acetate buffer equilibrium is shifted towards the basic. In TAP these two processes occur simultaneously resulting in a relatively stable pH, whereas in HSM only the acidifying uptake of NH₄+ occurs, resulting in a significant drop in the pH. In both cases, OD, pH, and measured temperature were shown to vary very little between replicates as shown in the figures 1, 2, and 3 respectively: error bars in each figure represent standard deviation.
C. reinhardtii
CC1609 was observed to reach stationary phase after around 3 days independent of growth medium; however, a higher final OD 740nm was achieved in TAP. Due to the economic benefits associated with HSM over TAP, the optimisation of C. reinhardtii
growth in HSM would be worth pursuing, with investigations into NO₃- as a nitrogen source as oppose to NH₄+ presenting the next logical step in this process.
References
Debuchy, R., Purton, S. and Rochaix, J.D. (1989) The argininosuccinate lyase gene of Chlamydomonas reinhardtii
: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. The EMBO journal
, 8 (10), p. 2803
Boynton, J.E., Gillham, N.W., Harris, E.H., Hosler, J.P. and Johnson, A.M. (1988) Chloroplast transformation in Chlamydomonas
with high velocity microprojectiles. Science
, 240 (4858), p.1534
Day, A. and Goldschmidt‐Clermont, M. (2011) The chloroplast transformation toolbox: selectable markers and marker removal. Plant biotechnology journal
, 9 (5), pp. 540-553
Harris, E.H. (2009) Chlamydomonas
Sourcebook: Introduction to Chlamydomonas
and Its Laboratory Use (Vol. 1), Academic Press
Kindle, K.L. (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii . Proceedings of the National Academy of Sciences
, 87 (3), pp. 1228-1232
Kumar, S.V., Misquitta, R.W., Reddy, V.S., Rao, B.J. and Rajam, M.V. (2004) Genetic transformation of the green alga- Chlamydomonas reinhardtii
by Agrobacterium tumefaciens. Plant Science
, 166 (3), pp.731-738
Mayfield, S.P., Manuell, A.L., Chen, S., Wu, J., Tran, M., Siefker, D., Muto, M. and Marin-Navarro, J., 2007. Chlamydomonas reinhardtii
chloroplasts as protein factories. Current opinion in biotechnology
, 18 (2), pp. 126-133
Merchant, S.S., Prochnik, S.E., Vallon, O., Harris, E.H., Karpowicz, S.J., Witman, G.B., Terry, A., Rochaix, J.D. (1995) Chlamydomonas reinhardtii
as the photosynthetic yeast. Annual review of genetics
, 29 (1), pp.209-230
Rosales-Mendoza, S., Paz-Maldonado, L.M.T. and Soria-Guerra, R.E. (2012) Chlamydomonas reinhardtii
as a viable platform for the production of recombinant proteins: current status and perspectives. Plant cell reports, 31(3), pp. 479-494
Scaife, M.A., Nguyen, G.T., Rico, J., Lambert, D., Helliwell, K.E. and Smith, A.G. (2015) Establishing Chlamydomonas reinhardtii
as an industrial biotechnology host. The Plant Journal
, 82 (3), pp. 532-546
Shimogawara, K., Fujiwara, S., Grossman, A. and Usuda, H. (1998) High-efficiency transformation of Chlamydomonas reinhardtii
by electroporation. Genetics
, 148 (4), pp. 1821-1828
Sodeinde, O.A. and Kindle, K.L. (1993) Homologous recombination in the nuclear genome of Chlamydomonas reinhardtii
. Proceedings of the National Academy of Sciences, 90 (19), pp.9199-9203
Torzillo, G., Scoma, A., Faraloni, C. and Giannelli, L. (2015) Advances in the biotechnology of hydrogen production with the microalga Chlamydomonas reinhardtii
. Critical reviews in biotechnology
, 35 (4), pp.485-496
PDF Version:
© 2024
Algenuity Limited
Registered in the United Kingdom No: 12456594
Site design by Ibby Tarafdar Creative
© 2024
Algenuity Limited. Registered in the United Kingdom No: 12456594
Site design by Ibby Tarafdar Creative